Introduction
Translation means decoding the mRNA and linking amino acids covalently together to form a polypeptide. In simple words, translation is the biological process in which messenger RNA is translated into proteins in prokaryotes.
This happens at the ribosome. Translation starts when a ribosome binds with the mRNA and is positioned such that the translation will give the correct amino acid sequence in the polypeptide chain. Transfer RNA takes amino acids to the ribosome. Then these amino acids can be added to the polypeptide chain as the ribosome moves down to the mRNA molecule.
Direction of protein syntesis
Protein synthesis also proceeds in one direction like DNA or RNA synthesis. Polypeptide formation starts with the amino acid at the end of the chain N-terminal with a free amino group and it will move in the C-terminal direction. Therefore, translation is said to happen in the amino terminus to the carboxyl terminus direction.
Efficiency of Translation Process In Bacteria
Protein synthesis is an accurate and rapid process. Cells with fast growth must use each mRNA efficiently to synthesize proteins at a rapid rate. For rapid rates of protein synthesis, mRNAs are simultaneously linked with several ribosomes forming a complex. Each ribosome will read the mRNA message and form a polypeptide. An mRNA complex with many ribosomes is called a polyribosome or polysome. Polysomes are present in all organisms.
Bacteria can also increase the efficiency of gene expression by doing transcription and translation simultaneously. As RNA polymerase synthesizes an mRNA, ribosomes already are attached to the mRNA so that transcription and translation happen simultaneously. Simultaneous transcription and translation are possible in bacterial cells because the nuclear envelope does not separate the translation machinery from DNA, unlike in eukaryotes.
Amino Acid Activation
Amino acid activation involves the attachment of an Amino Acid to a transfer RNA. For the start of translation, a supply of tRNA molecules must be ready that have the correct amino acid. So, there is a preparatory step for protein synthesis called as amino acid activation. In this process, amino acids are attached to tRNA molecules.
Structure of tRNA molecules
Transfer RNA molecules (tRNA) are about 70 to 95 nucleotides long and they have many characteristic structural features. These features become clear when the tRNA is folded and the base pairing within the tRNA strand is maximized. The two-dimensional representation shows that this base pairing causes the tRNA to look like a cloverleaf conformation and the three-dimensional representation shows that the structure looks like the letter L.
Features of Transfer Rna
The acceptor stem is one important feature of tRNAs. It holds the activated amino acid. The 3′ ends of all tRNA molecules have the same CCA sequence and in all cases, the amino acid is attached to the 3′-hydroxyl of the adenosine nucleotide.
Anticodon is another important feature of a tRNA. The anticodon is complementary to an mRNA codon. It is located on the anticodon arm. Aminoacyl-tRNA synthetases are the enzymes that catalyze amino acid activation. The attachment of amino acid to the tRNA is a high-energy bond. In this bond, energy is stored that provides the fuel which is needed to generate the peptide bond when the amino acid is added to the growing peptide chain.
Accuracy in the translation process
There are 20 aminoacyl-tRNA synthetases at least and each of them is specific for a single amino acid and its tRNAs. It is important that each tRNA is attached to the corresponding amino acid because if an incorrect amino acid gets attached to the tRNA, it will be incorporated into a polypeptide in place of the correct amino acid. The synthetic protein machinery can only recognize the anticodon of the aminoacyl-tRNA and cannot differentiate whether the correct amino acid is attached or not. There are some aminoacyl-tRNA synthetases that proofread just like the DNA polymerases. If the wrong amino acid gets attached to the tRNA, the enzyme will hydrolyze the amino acid from the tRNA rather than release the incorrect product.
Transfer RNA binding sites
There are three binding sites of tRNA. Protein synthesis occurs in the ribosomes. Ribosomes have two subunits; the large subunit and the small subunit. Each subunit contains one or more rRNA molecules and many proteins. The ribosomes are divided into two functional domains, first is the translational domain and the second is exit domain. Both subunits help in the formation of the translational domain that interacts with tRNAs and because of this peptide bonds are formed. The exit domain is present only in the large subunit.
There are three sites in the translational domain for the binding of tRNAs: A, P, and E sites. The A site called aminoacyl or acceptor site receives tRNAs that carry an amino acid which is to be added to the protein being formed. The P site knowns as the peptidyl or donor site hold the tRNA that is attached to the growing polypeptide. The E site called as exit site is the place from where empty tRNAs leave the ribosome. The emergence of a growing peptide chain occurs from the large subunit at the exit domain.
Role of ribosomal RNA
Ribosomal RNA rRNA has three roles:
- All three rRNA molecules help in the structure of ribosomes.
- For initiation of protein synthesis, the 16S rRNA of the 30S subunit is needed because its 3′ end attaches to a site on the leader of the mRNA called the Shine-Dalgarno sequence. It means that the Shine-Dalgarno sequence is part of the ribosome-binding site (RBS). This will position the mRNA on the ribosome. The 16S rRNA also attaches to a protein that is needed to start the translation (initiation factor 3) and also to the 3′ CCA end of aminoacyl-tRNA.
- The peptide bond formation is catalyzed by the 23S rRNA which is a ribozyme.
Protein synthesis
Protein synthesis is divided into three stages: initiation, elongation, and termination.
Initiation
Bacterial protein synthesis starts with a modified molecule called aminoacyl-tRNA, N-formyl methionyl-tRNA fMet (fMet-tRNA). It is also known as the initiator tRNA and is coded for the start codon AUG. The initiator tRNA molecules have a formyl group that is covalently bound to the amino group and it can be used only for the initiation. When methionine is need to be added to a growing polypeptide chain such that at an AUG codon in the middle of the mRNA; a normal methionyl-tRNAMet is employed. Although bacteria begin their protein synthesis with N-formyl-methionine. The formyl group is not retained and it is removed hydrolytically.
30S Initiation complex formation
30S initiation complex consists of the initiator tRNA, mRNA that is to be translated, the 30S ribosomal subunit, and two initiation factors (IF-1 and IF-2). Initiator fMet-tRNA position on the mRNA is very important for proper translation of the mRNA and this happens with the help of the 16S rRNA within the 30S subunit. The start codon (AUG) specifically attaches with the fMet-tRNA anticodon by aligning the Shine-Dalgarno sequence with the 16S rRNA. This will ensure that the start codon may translate first.
Formation of 70S Initiation complex
Protein synthesis starts with the formation of the 70S initiation complex just like transcription and DNA replication. When the 30S initiation complex forms, it will bind to the 50S ribosomal subunit that will form the 70S initiation complex. The fMet-tRNA will be placed at the peptidyl or P site. At this point, the initiation factor (IF-3) will keep the binding of 30S and 50S subunits to each other.
Energy for initiation
GTP is a high-energy molecule like ATP. Hydrolysis occurs which covert GTP to GDP that provides the energy needed for the initiation.
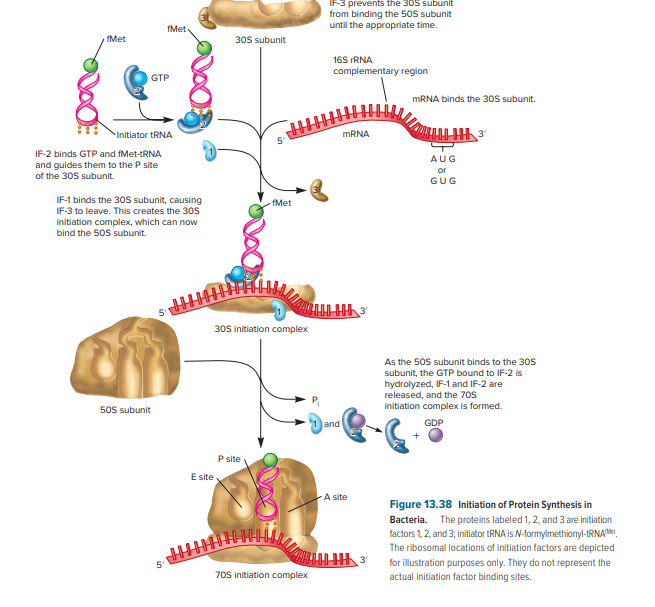
Figure 1: This figure represents the initiation stage of translation in prokaryotes. Firstly, 30S initiation complex is formed then the 70S initiation complex formation occurs.
Elongation
The addition of an amino acid to a growing polypeptide chain is the result of an elongation cycle that is composed of three phases: aminoacyl-tRNA binding, transpeptidation reaction, and translocation. The process is helped by proteins called elongation factors (EF).
An amino acid corresponds to the proper mRNA codon and is added to the C-terminal end of the polypeptide chain by the movement of ribosome down the mRNA in the direction 5 ′ to 3′, in each turn of the cycle. At the start of an elongation cycle, the P site is filled with the initiator fMet-tRNA. A and E sites are empty. Messenger RNA (mRNA) is attached to the ribosome in such a way that only the proper or accurate codon interacts with the P site tRNA (i.e. an AUG codon for fMet-tRNA). The next codon will be located at the A site and will be ready to receive an aminoacyl-tRNA.
Amino acyl-tRNA Binding phase
the first phase of the cycle is an aminoacyl-tRNA binding phase. In this phase of aminoacyl-tRNA binding, the aminoacyl-tRNA that corresponds to the codon located in the A site is inserted so that its anticodon will align with the codon on the mRNA. In the case of bacterial cells, this process occurs with the help of two elongation factors and needs energy in the form of one GTP. When the proper aminoacyl-tRNA is placed in the A site, then in the second phase of the elongation cycle, the transpeptidation reaction starts.
Transpeptidation reaction
The transpeptidation reaction is catalyzed by the enzyme known as a peptidyl transferase. This enzyme is of 23S rRNA ribozyme, which is the part of 50S ribosomal subunit. The transpeptidation reaction involves the reaction between the amino group of the A-site amino acid with the carboxyl group of the C-terminal amino acid at the P site tRNA. Because of this reaction transfer of the peptide chain occurs from the tRNA in the P site to the tRNA in the A site due to peptide bond formation between the peptide chain and the incoming amino acid. There is no need for an extra energy source for peptide bond formation because the bond linkage of an amino acid to tRNA is already high in energy.
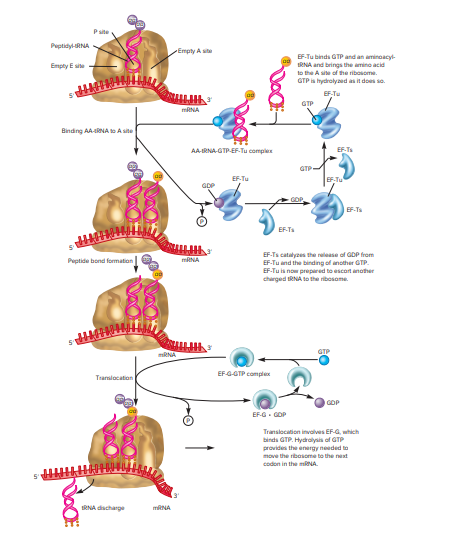
Figure 2: This figure represents the elongation stage of translation in bacteria. The function of elongation factors and the movement of ribosomes is illustrated.
Translocation
Translocation is the final phase in the elongation cycle. In this phase, three things happen simultaneously:
- the peptidyl-tRNA will move from the A site to the P site
- the ribosome will move one codon along the mRNA so that a new codon will be positioned in the A site
- the empty tRNA will move from the P site to the E site and will leave the ribosome.
Translocation includes the 30S and 50S subunit’s rotations relative to each other. Additionally, the head portion of the 30S subunit turns around. These changes in the structure of ribosomes move the tRNAs into their new locations. The interactions of codon and anticodon between the tRNAs and the mRNA move the mRNA with the movement of tRNA. One elongation factor will participate and one GTP will be hydrolyzed during this complex process.
Termination
The end of protein synthesis occurs when the ribosome will reach a Stop Codon. There are three stop codons: UAA, UAG, and UGA. The stop codon is found on the mRNA. There are three release factors; RF-1, RF-2, and RF-3 that help the ribosome in recognizing these codons. As there is no related tRNA for a stop codon, the ribosome will stop. Peptidyl transferase will hydrolyze the bond linkage of the polypeptide to the tRNA in the P site. Then the polypeptide and the empty tRNA will release. The ribosome recycling factor is a protein that structurally mimics the tRNA. This protein is responsible for the disassembling of the translational complex. The mRNA will release and the two ribosomal subunits will get separated. IF-3 will bind to the 30S subunit, which will prepare it for the next round of protein synthesis.
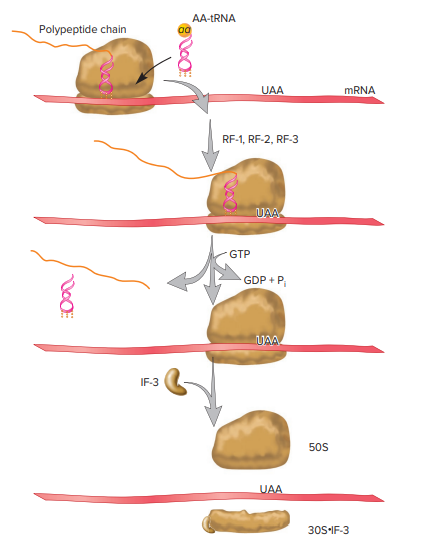
Figure 3: This figure represents the termination of translation. UAA has mostly used end codons as shown.
REFERENCES
Willey, J. M., Sandman, K. M., Wood, D. H., & Prescott, L. M. (2019). Prescott’s microbiology (11th ed.). McGraw Hill.