DNA replication in eukaryotes
The word eukaryotes is a composite of two greek words i.e., EU which means well or good, and karyon which means nut or kernel. Eukaryotes are those organisms that have a visible nucleus enclosed in a nuclear envelope. Eukaryotes are one of the three domains of life. Organisms such as animals, plants, fungi, and protists are examples of eukaryotes.
DNA replication
It is a process in which DNA makes a copy of itself before cell division in order to pass the accurate amount of information to the daughter cells. DNA replication in eukaryotes follows a semi-conservative model, this means the two old DNA strands serve as a template for the formation of an entirely new strand. Because each of the two daughters of a dividing cell inherits a new DNA double helix containing one old and one new strand.
Although the eukaryotic system is significantly more complex in terms of the amount of DNA to be copied and the number of proteins required, the mechanisms for eukaryotic and prokaryotic DNA replication are quite similar.
Enzymes involved in eukaryotic DNA replication
Polymerases α, δ, and ε, which are all members of the b-family of polymerase, are the three major enzymes responsible for replicating eukaryotic nuclear DNA.
Polymerase α:
Like all DNA polymerases, DNA polymerase α (pol α) replicates DNA by extending a primer in the 5’ → 3’ direction while being guided by a single stranded DNA template. Since this enzyme lacks exonuclease activity, it cannot edit the polymerization product.
Pol α is firmly associated with a primase and only moderately processive (polymerize 100 nucleotides at a time), suggesting that it is engaged in the start of DNA replication in eukaryotes. A 7 to 10 nucleotide RNA primer is created by the pol α–primase complex, and it is then extended by about 15 deoxynucleotides. Since the RNA primer and the initial few residues of freshly produced DNA are frequently deleted and replaced, its lack of proofreading activity is not problematic.
Polymerase δ:
The active site of the 3’ to 5’ exonuclease is present in DNA polymerase δ (pol δ), which does not bind with a primase. Furthermore, pol δ has a virtually infinite response (it can replicate the entire template DNA), but only when it is in association with the sliding-clamp protein known as proliferating cell nuclear antigen (PCNA). PCNA forms a trimeric ring with almost exactly the same structure (and probably function) as the e. Coli β2 sliding clamp, according to kuriyan’s analysis of its x-ray structure. Interestingly, despite the alignment of their physically related parts, PCNA and the clamp show no distinct sequence identity.
DNA synthesis requires PCNA and pol δ in a complex. The clamp loader RFC, which is the eukaryotic equivalent of the e. Coli γ complex loads PCNA onto the template strand close to the primer during replication. This pushes pol α out of the way, allowing pol δ to attach to and gradually lengthen the new DNA strand.
Polymerase ε:
In the absence of PCNA, DNA polymerase ε (pol ε) mimics pol δ on the surface but is much more processive. It has been suggested that pol ε serves as the leading-strand polymerase while pol α and pol δ work together to manufacture the lagging strand because of it and it is necessary for DNA replication in eukaryotes.
Some other enzymes are also involved in the eukaryotic DNA replication
Helicases
It unwinds the double helix of DNA.
Topoisomerases
It is involved in the removal of supercoiling.
Ligase
It helps in the joining of fragments.
Steps of DNA replication in eukaryotes
- Initiation
- Elongation
- Termination
Initiation
The first stage of DNA synthesis is where the DNA double helix unwinds and a replication fork forms at the origin of replication.
Pre-initiation:
Before initiation, DNA is made accessible to proteins and enzymes. DNA replication in eukaryotes starts from a sequence known as the origin of replication (ORI) also known as replisomes. Eukaryotic cells have multiple sites for replication to initiate. To initiate, various replicative proteins must assemble on these sites.
Proteins involved in initiation:
- Origin replication complex (ORC): it has ATPase activity and recruits other proteins
- Cell division cycle 6 (CDC 6): it helps in helicase loading
- Cdc 10 – dependent transcript 1(CDC-1): it also helps in mcm/helicase loading
- Minichromosomal maintenance (mcm-2-7): it has helicase activity
- Cyclin dependent kinase (CDK): it is involved in the initiation of replication proteins by phosphorylation
- Cell division cycle 45 (CDC-45): it interacts with mcm 7 and polymerase alpha
- Gins: it aids in elongation
- Replication protein A (RPA): it prevents reannealing of DNA strands and ssb’s (single stranded DNA binding proteins)
- Replication factor c (RFC): it acts as a DNA clamp loader and loads PCNA
- Proliferating cell nuclear antigens (PCNA): it acts as DNA sliding clamp (holds DNA)
Formation of the pre-replication complex:
A pre-replicative complex (pre-RC) is formed in the following steps:
- Association of orc occurs with ORI then binding of cdc6 happens and then CDTI & MCM binds.
- Orc assembles during the G1 phase before the S phase, and CDK and DDK proteins bind to the orc.
- This transforms pre-RC into an active replication fork.
- Certain proteins recognize ORI and allow other DNA replication proteins to bind to the same region which stimulates other DNA replication proteins to bind.
- Among these proteins is the helicase enzyme, which unwinds and separates the DNA helix into single stranded DNA.
- As DNA opens up, Y-shaped structures called “Replication forks” are formed at the replication site.
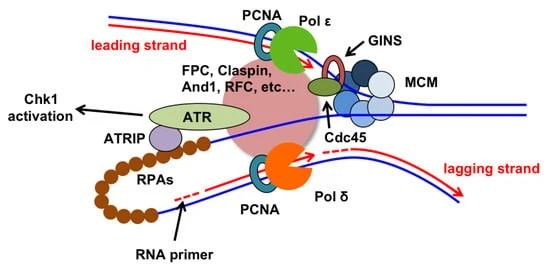
Formation of replication fork:
The Replication fork consists of 4 components:
- DNA helicase and DNA polymerase δ (they both are associated with helicase activity).
- DNA polymerase α involved in the synthesis of RNA primers (DNA polymerase α has primer activity).
- DNA polymerase ε is involved in the lagging strand and DNA polymerase δ in the leading strand initiates daughter strand synthesis.
- Ssb (single-stranded DNA binding protein) and RPA bind to ss DNA and prevent re-annealing of the ss DNA
Multiple origins of replication on the eukaryotic chromosome allow replication to occur simultaneously in hundreds to thousands of locations along each chromosome.
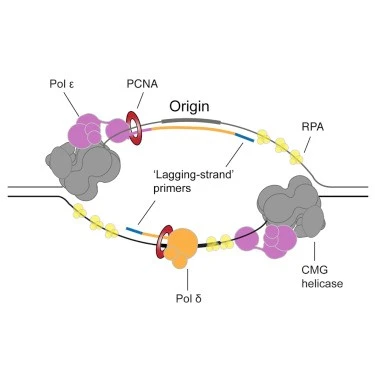
Elongation
DNA polymerase adds DNA nucleotides to the 3′ ends of the newly synthesized polynucleotide strand. The template strand specifies which of the four DNA nucleotides (a, t, c, or g) is added at each position along the new chain. DNA polymerase contains a groove that allows it to bind to a single-stranded template DNA and travel one nucleotide at a time. DNA polymerase cannot initiate new strand synthesis; it only adds new nucleotides at the 3′ ends of an existing strand. All newly synthesized polynucleotide strands must be initiated by a specialized RNA polymerase called primase.
Primase initiates polynucleotide synthesis by creating a short RNA polynucleotide strand complementary to the template DNA strand. This short stretch of RNA nucleotides is called the primer. Once RNA primer has been synthesized at the template DNA, primase exits, and DNA polymerase extends the new strand with nucleotides complementary to the template DNA. Eventually, the RNA nucleotides in the primer are removed and replaced with DNA nucleotides. Once DNA replication in eukaryotes is finished, the daughter molecules are made entirely of continuous DNA nucleotides, with no RNA portions.
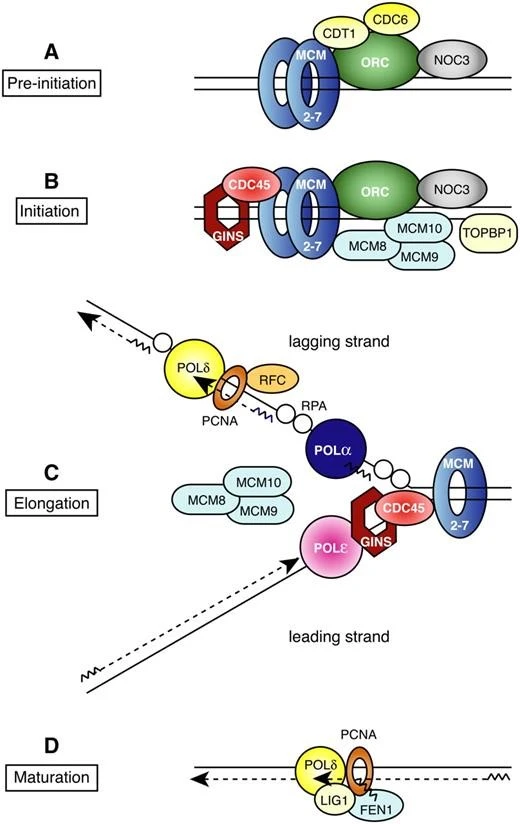
The leading and lagging strands:
As DNA polymerase synthesizes new strands in the 5’ to 3’ direction, two newly synthesized strands grow in opposite directions because template strands at each replication fork are anti parallel. The “leading strand” is synthesized continuously towards the replication fork as helicase unwinds the template double-stranded DNA. The “lagging strand” is synthesized in the direction away from the replication fork and away from the DNA helicase unwinds. This lagging strand is synthesized in pieces because the DNA polymerase can only synthesize in the 5′ to 3′ direction, and so it constantly encounters the previously-synthesized new strand. The pieces are called Okazaki fragments, and each fragment begins with its own RNA primer.
Termination
Replication is terminated when two replication forks counter each other
- Scf e3 (ubiquitin ligase complex) comes in and catalyzes mcm protein
- P97 is the segregate protein that segregates and inactivates mcm
DNA polymerase can’t form a phosphodiester bond between two segments of new DNA strands, these unattached segments are called “nicks”. Rna primers need to be replaced with DNA and nicks in the sugar-phosphate backbone need to be connected.
Flap endonuclease 1 (fen1) and RNase h remove RNA primers at the start of each leading strand and at the start of the Okazaki fragment, leaving gaps of un-replicated template DNA. A free-floating DNA polymerase then binds at the 3’ end and extends the DNA over the gap. Finally, ligase joins the sugar phosphate backbones at each nick site. After connecting all nicks, the new strand is one long continuous DNA strand, and the daughter DNA molecule is complete.
Problems in the completion of lagging strand:
Removal of RNA primer from the 5’ end of a linear molecule would leave a gap i.e., primer gap. The gap cannot be filled by DNA polymerase action, because of the absence of a primer terminus to extend. This leads to the shortening of the daughter strand a bit with each round of replication. This problem is solved by telomerase.
Telomerase
It is a ribonucleoprotein, containing RNA components 9-30 nucleotides long. This RNA component serves as a template for the synthesis of telomeric repeats at the parental DNA ends. Telomerase is RNA dependent DNA polymerase with RNA components. It uses:
- 3’ end of parental DNA as a primerRna component of telomerase as a
- template
- Adds successive telomeric repeats to the parental DNA strand at its 3’ ends
Features of eukaryotic DNA replication:
- Replication is bi-directional and originates at multiple origins of replication (ori c) in eukaryotes.
- It occurs only in the s phase and at many chromosomal origins.
- It takes place in the cell’s nucleus.
- Synthesis occurs only in the 5′ to 3′direction.
- Strands of DNA are manufactured in different directions, producing a leading and a lagging strand.
- Eukaryotic cells possess five types of polymerases involved in the replication process
References:
- Aria V, Yeeles JTJMc. 2019. Mechanism of bidirectional leading-strand synthesis establishment at eukaryotic DNA replication origins. 73(2): 199-211. e110.
- Leman AR, Noguchi EJG. 2013. The replication fork: understanding the eukaryotic replication machinery and the challenges to genome duplication. 4(1): 1-32.
- Shultz RW, Tatineni VM, Hanley-Bowdoin L, Thompson WFJPP. 2007. Genome-wide analysis of the core DNA replication machinery in the higher plants Arabidopsis and rice. 144(4): 1697-1714.
- Eukaryotic_DNA_replication