Introduction
Overview
A Gram-negative bacterium that is a member of the Pseudomonadaceae family, Pseudomonas aeruginosa is very important in microbiology, especially when it comes to isolation and identification in environmental and clinical situations. P. aeruginosa is a ubiquitous organism that demonstrates exceptional flexibility, flourishing in a variety of settings, including soil, water bodies, and medical contexts. Because of its inherent resistance mechanisms and frequent occurrence in healthcare settings, it is a primary cause of nosocomial infections, necessitating strict isolation and identification procedures.
For the purpose of comprehending P. aeruginosa’s pathogenicity, antibiotic resistance profiles, and epidemiology, it is essential to isolate and correctly identify the bacteria. The distinct features of this bacterium, such as its rod-shaped morphology, motility, and capacity to synthesize distinctive pigments like pyocyanin, make specialized methods necessary for efficient isolation and identification. Moreover, improved sensitivity and specificity in identifying P. aeruginosa strains are provided by molecular methods that target certain genes, including the oprL gene.
This article explores the complexities involved in isolating and identifying Pseudomonas aeruginosa, delving into sophisticated molecular techniques, biochemical assays, and conventional culture-based approaches.
Isolation Techniques
Sample Collection
Types of Samples
- Water: Pseudomonas aeruginosa is frequently found in water sources, particularly in contaminated areas. A few possible sites for sampling are rivers, lakes, and sources of drinking water.
- Clinical Specimens: Sputum, urine, and wound swabs from individuals who may have had infections with P. aeruginosa. These samples are essential for both identifying and tracking infections brought on by this microorganism.
- Environmental Samples: P. aeruginosa may be present in a variety of environmental samples, such as plants, soil, and surfaces of medical equipment. These samples also have the potential to act as reservoirs for transmission.
Enrichment Media
Because enrichment media offer a nutrient-rich environment that is favourable to Pseudomonas aeruginosa growth, they are essential in the isolation of the bacterium from complex samples. P. aeruginosa enrichment medium that are frequently utilized include:
- Pseudomonas Selective Broth (PSB)
- Cetrimide Broth
- King’s B Medium
- Nutrient Broth Enhanced with Certain Supplements
Particular Supplements for Nutrient broth:
- 4% Sodium Chloride (NaCl)
- 0.05% Crystal Violet
These broth media have specific chemicals in them that prevent competing microbes from growing while promoting the growth and multiplication of P. aeruginosa. Following the samples’ inoculation into the enrichment broth, P. aeruginosa grows when the right circumstances are met during incubation, resulting in the development of a culture that is enriched with the target organism.
Isolation on Solid Media
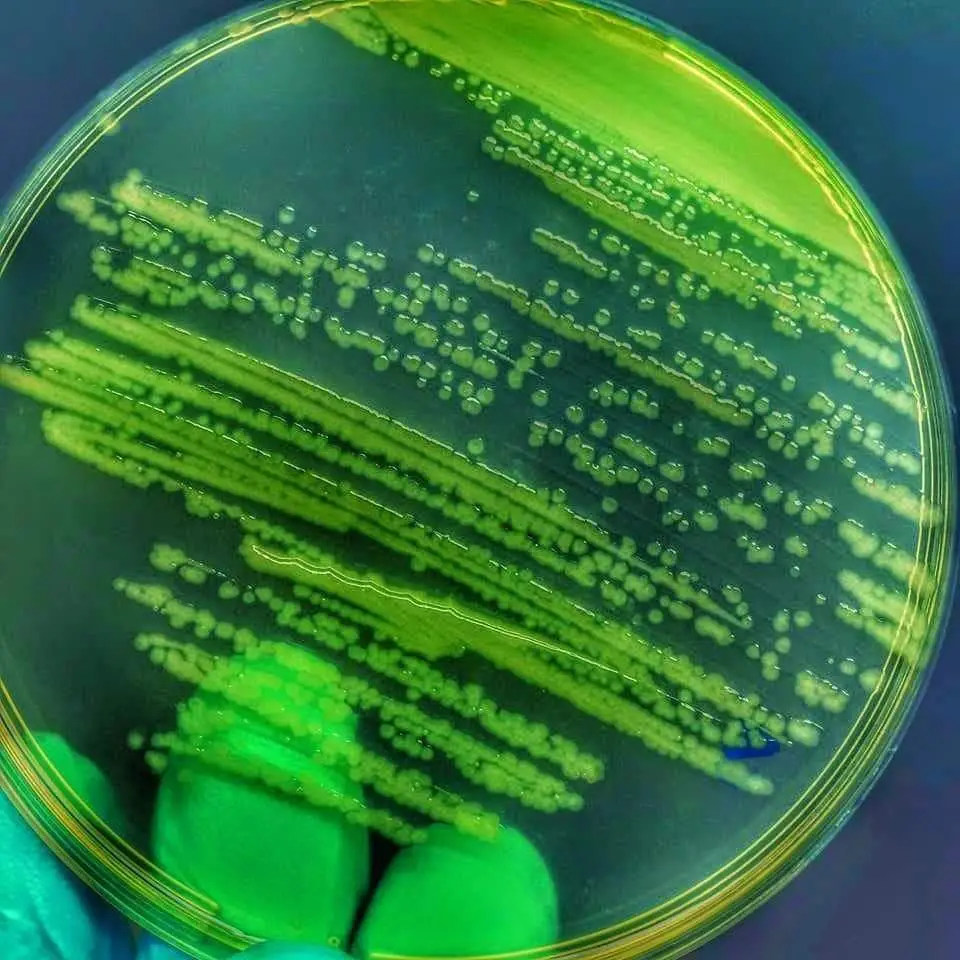
1. Nutrient Agar
On nutrient agar, Pseudomonas aeruginosa displays distinctive colony morphology and growth patterns specific to its characteristics:
Colony Morphology
- Color: Bluish-green or greenish-blue colonies.
- Appearance: Irregular, spreading colonies with a mucoid or slimy texture.
- Size: Medium to large-sized colonies.
- Texture: Slimy or viscous appearance due to the production of extracellular polysaccharides.
Growth Patterns
- Growth Rate: Moderate to fast growth. Pseudomonas aeruginosa typically exhibits rapid growth on nutrient agar, forming colonies within 24-48 hours.
- Colonial Characteristics: Individual colonies may appear rough or wrinkled, with irregular edges and a shiny surface.
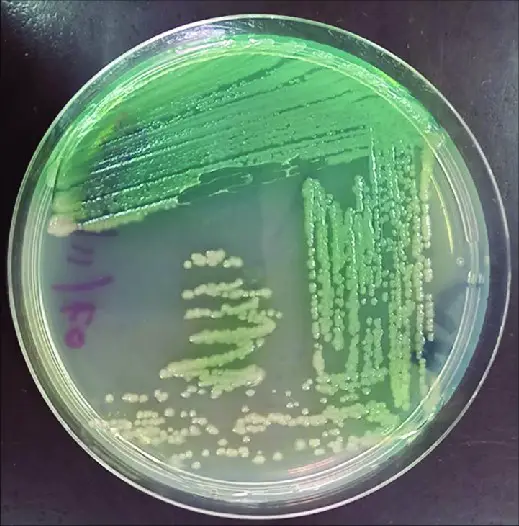
2. Pseudomonas Agar
On Pseudomonas agar, Pseudomonas aeruginosa demonstrates specific colony morphology and growth patterns ideal for its isolation and identification:
Colony Morphology
- Color: Bluish-green or greenish-blue colonies, similar to those on nutrient agar.
- Appearance: Slimy or mucoid colonies with irregular margins.
- Size: Variable, ranging from small to large-sized colonies.
- Texture: Slimy or viscous appearance due to the production of alginate and other extracellular polysaccharides.
Growth Patterns
- Pyocyanin Production: Positive. Pseudomonas aeruginosa produces the characteristic blue-green pigment pyocyanin, imparting color to the colonies.
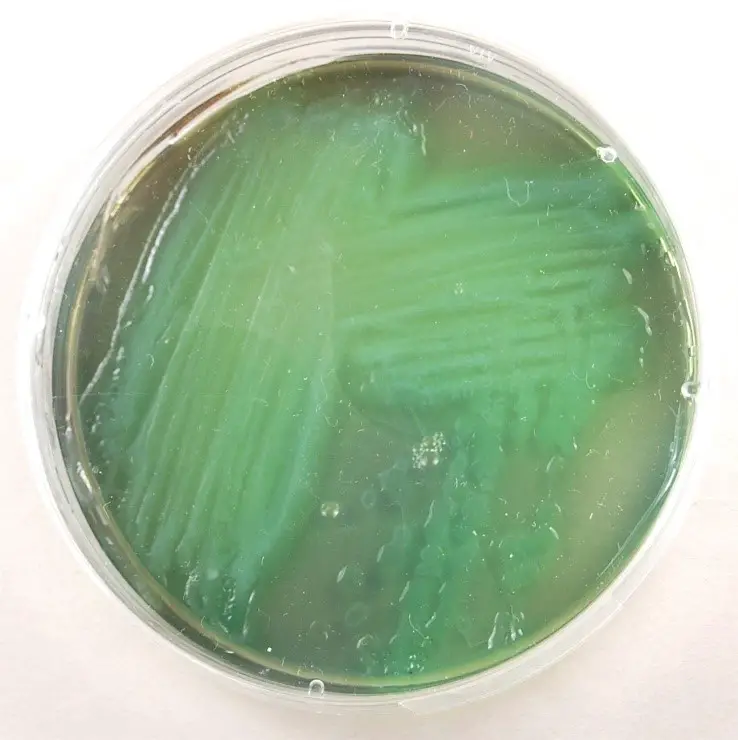
3. Cetrimide Agar
Cetrimide agar is a selective medium designed to isolate Pseudomonas species, including Pseudomonas aeruginosa, from mixed cultures:
Colony Morphology
- Color: Bluish-green or greenish-blue colonies, similar to those on Pseudomonas agar.
- Appearance: Smooth or slightly mucoid colonies.
- Size: Variable, ranging from small to large-sized colonies.
- Texture: Glistening or shiny appearance. Growth Patterns:
- Selective Inhibition: Cetrimide in the agar selectively inhibits the growth of other bacterial species, allowing for the selective isolation of Pseudomonas aeruginosa.
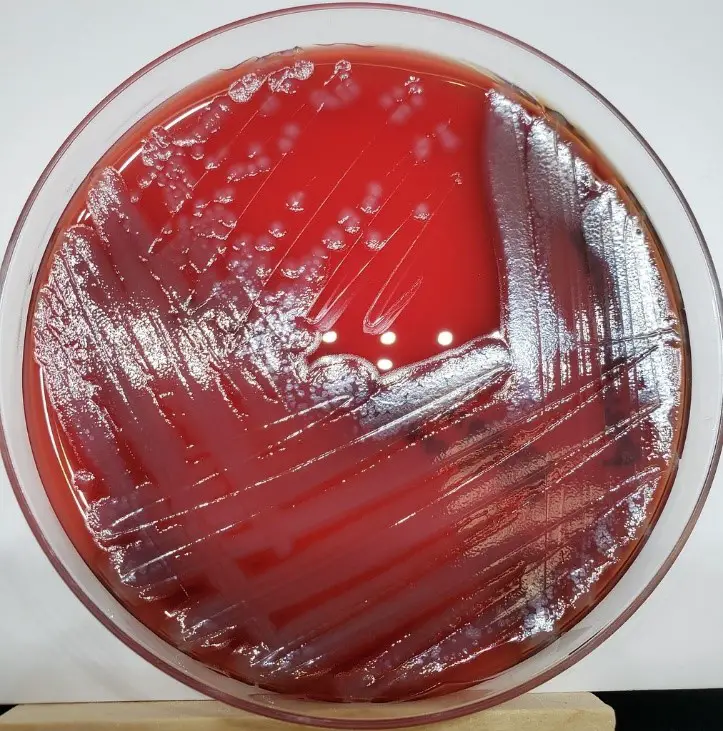
4. Blood Agar
Blood agar is a non-selective medium commonly used for the cultivation of a wide range of microorganisms, including Pseudomonas aeruginosa for the observation of hemolytic activity.
Colony Morphology
- Color: colonies appear gray/gray-white with a yellowish tint and exhibit a silvery sheen.
- Appearance: Smooth or mucoid colonies.
- Size: Medium-sized colonies.
- Texture: Glistening or moist appearance.
Growth Patterns
- Beta-hemolysis: Complete lysis of red blood cells around the colonies, indicating beta-hemolytic activity.
5. Eosin Methylene Blue (EMB) Agar
Eosin Methylene Blue (EMB) agar is selective for gram negative bacteria therefore supporting the growth of pseudomonas aeruginosa and inhibits the growth of gram-positive bacteria.
Colony Morphology
- Color: Typically, Pseudomonas aeruginosa colonies on EMB agar appear as pale or light pink with a metallic sheen.
- Appearance: Smooth or mucoid colonies.
- Size: Medium-sized colonies.
- Texture: Glistening or moist appearance.
Growth Patterns
- Lactose Fermentation: Negative. Pseudomonas aeruginosa does not ferment lactose, so there is no color change in the colonies or the surrounding medium.
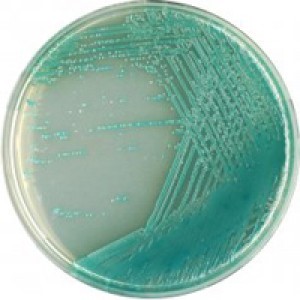
6. Chromogenic Agar
Chromogenic agar, such as Chrom agar, is a type of selective and differential medium that contains chromogenic substrates to aid in the identification of specific bacterial species based on the color of their colonies
Colony Morphology
- Color: Colonies of Pseudomonas aeruginosa on chromogenic agar typically appear as greenish blue.
- Appearance: Smooth or mucoid colonies.
- Size: Small to medium-sized colonies.
- Texture: Moist appearance.
Growth Patterns:
- Chromogenic Substrate Utilization: The green color of colonies is indicative of pyocyanin production by Pseudomonas aeruginosa.
Microscopic Examination
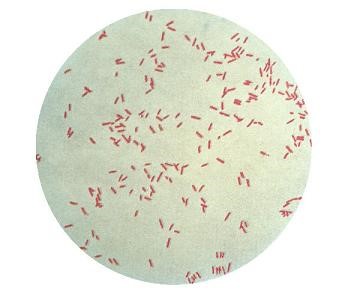
1. Gram Staining
After Gram staining, pseudomonas aeruginosa, a Gram-negative bacterium, looks pink or red under a microscope.
2. Morphological Features Observed Under a Microscope
- The morphology of pseudomonas aeruginosa usually manifests as tiny, slender rods or bacilli. Cells can be found single, in pairs, or in chains.
- Pseudomonas aeruginosa may or may not exhibit motility because to the presence of polar flagella in some strains while absent in other strains therefore motility varies in different strains of organism, which can be seen using the hanging drop method.
- Cell size can be estimated through microscopic analysis; for Pseudomonas aeruginosa, this typically ranges from 0.5 to 1 micrometres in diameter and 1 to 3 micrometres in width.
Biochemical Tests
1. SIM Test
The SIM (Sulfide, Indole, Motility) test assesses key characteristics of Pseudomonas aeruginosa:
- Sulfide Production: Negative (no blackening of the medium).
- Indole Production: Typically negative, as Pseudomonas aeruginosa does not produce indole.
- Motility: Variable, as some strains may exhibit motility, while others are non-motile.
2. Oxidase Test
- P. aeruginosa is typically oxidase-positive, exhibiting a color change (usually blue) upon exposure to oxidase reagents.
- This test is crucial for differentiating P. aeruginosa from other Gram-negative bacteria.
3. Catalase Test
- P. aeruginosa is catalase-positive, producing rapid bubbles upon exposure to hydrogen peroxide.
- This test helps in distinguishing P. aeruginosa from other catalase-negative bacteria.
4. Methyl Red (MR) Test
- P. aeruginosa typically yields a negative MR test result due to its preference for oxidative metabolism, resulting in a lack of significant acidic end-product formation.
- This test aids in differentiating P. aeruginosa from MR-positive organisms.
5. Voges-Proskauer (VP) Test
- The VP test result for Pseudomonas aeruginosa is typically negative, as it predominantly follows oxidative metabolic pathways.
6. Triple Sugar Iron (TSI) Agar Test
- P. aeruginosa typically produces alkaline slant and acidic butt in TSI agar, resulting in a yellow slant and red butt.
- Gas formation may be observed, indicated by cracks in the media or the media raised from the bottom of the test tube.
- This test helps in confirming the identity of P. aeruginosa and assessing its sugar fermentation patterns.
Biochemical profile of P. Aeruginosa
| Basic Characteristics | Properties (P. aeruginosa) |
| Gram Staining | Negative |
| Shape | Rods (bacilli) |
| Motility | Varies in strains – Motile (Unipolar) |
| Flagella | Single Flagella |
| Capsule | Non-Capsulated |
| Spore | Non-Spore forming |
| Oxidase | Positive (+ve) |
| Catalase | Positive (+ve) |
| Coagulase | Negative (-ve) |
| Indole | Negative (-ve) |
| Citrate | Positive (+ve) |
| Urease | Negative (-ve) |
| Cetrimide Test | Positive (+ve) |
| MR | Negative (-ve) |
| VP | Negative (-ve) |
| OF (Oxidative/Fermentative) | Oxidative |
| Nitrate Reduction | Positive (+ve) |
| H2S | Negative (-ve) |
| Gas | Positive (+ve)- From Nitrate |
| Pigment | Positive (+ve) – Blue/Green |
| TSI Test | Alkali/Alkali (Red slant/Red butt) |
| Gelatin Hydrolysis | Positive (+ve) |
| Hemolysis | Beta hemolysis |
Sugar Fermentation profile of P. Aeruginosa
| Sugars | Fermentation Reactions |
| Fructose | Positive (+ve) |
| Malonate | Positive (+ve) |
| Mannitol | Positive (+ve) |
| Ribose | Positive (+ve) |
| Sucrose | Negative (-ve) |
| Lactose | Negative (-ve) |
| Galactose | Negative (-ve) |
| Maltose | Negative (-ve) |
| Glucose | Negative (-ve) |
| Mannose | Negative (-ve) |
| DNase | Negative (-ve) |
| Cellobiose | Negative (-ve) |
| Starch | Negative (-ve) |
| Hippurate | Negative (-ve) |
| Arabinose | Negative (-ve) |
| Sorbitol | Negative (-ve) |
| Xylose | Negative (-ve) |
| Trehalose | Negative (-ve) |
| Inositol | Negative (-ve) |
| Adonitol | Negative (-ve) |
| Inulin | Negative (-ve) |
| MyoInositol | Negative (-ve) |
Enzymatic reactions profile of E. coli
| Enzymes | Enzymatic Reactions |
| Acetate Utilization | Positive (+ve) |
| Arginine dehydrolase | Positive (+ve) |
| Lipase | Positive (+ve) |
| Acid Phosphatase | – |
| Beta Lactamase | – |
| Acetoin Production | – |
| Alkaline Phosphatase | – |
| Perooxidase | – |
| Hyalurodinase | – |
| Ornithine decarboxylase | Negative (-ve) |
| Lysine | Negative (-ve) |
| Lecithinase | Negative (-ve) |
| Phenylalanine deaminase | Negative (-ve) |
Molecular Techniques for Pseudomonas aeruginosa Identification
Polymerase Chain Reaction (PCR)
For the accurate identification of Pseudomonas aeruginosa, polymerase chain reaction (PCR) is a useful technique as it amplifies the specific portions of genes necessary for bacterial identification.
Target Genes for P. aeruginosa Identification
In PCR-based tests, a number of target genes are used for the identification and verification of P. aeruginosa:
- oprL Gene: When identifying P. aeruginosa, the oprL gene—which codes for the primary outer membrane protein OprL—is usually the first place to look. This gene, which is crucial for preserving antibiotic resistance and cell integrity in Pseudomonas species, is highly conserved.
- rpoD Gene (RNA Polymerase Sigma Factor): Another important target for P. aeruginosa identification is the rpoD gene, which codes for the RNA polymerase sigma factor. This gene is necessary for the development and survival of bacteria and is involved in the regulation of gene expression.
- 16S-23S rRNA Intergenic Spacer (ITS) Region: Variable sequences can be targeted in the intergenic spacer region between the 16S and 23S rRNA genes to identify P. aeruginosa species specifically. This region can be amplified using PCR to distinguish P. aeruginosa from other closely related species.
- exoA Gene (Exotoxin A): P. aeruginosa-specific exoA gene, which codes for the powerful virulence factor Exotoxin A, is frequently the target of PCR tests used to identify pathogenic strains. The identification of the exoA gene confirms the existence of P. aeruginosa strains that may be pathogenic.
These target genes enable the precise and quick molecular identification of Pseudomonas aeruginosa in clinical and environmental samples, when combined with suitable primer design and PCR amplification procedures.
DNA Sequencing
P. aeruginosa’s whole genome can be analyzed or certain genetic areas can be studied using DNA sequencing techniques, including Sanger sequencing and next-generation sequencing (NGS). P. aeruginosa strains can be precisely identified and described by comparing sequencing data to reference sequences or databases in NCBI.
FISH, or Fluorescence In Situ Hybridization
Fluorescence-labeled probes are used in FISH, and they hybridise to complementary regions in the P. aeruginosa genome. Using fluorescent microscopy, this method makes P. aeruginosa cells visible and identifiable in clinical or environmental samples.
References
- Brooks, T., Keevil, C. W., & MacDonald, N. (1997). A simple method for the rapid identification of Pseudomonas aeruginosa. Journal of Microbiological Methods, 29(3), 197-200.
- Janda, J. M., & Abbott, S. L. (2010). The genus Pseudomonas. In Manual of Clinical Microbiology (10th ed., pp. 719-729). American Society of Microbiology.
- von Gotz, F., Haussler, S., Jordan, D., Saravanamuthu, S. S., Wehmhoner, D., Strussmann, A., & Steinmetz, I. (2004). Expression analysis of a highly adherent and cytotoxic small colony variant of Pseudomonas aeruginosa isolated from a lung of a patient with cystic fibrosis. Journal of Bacteriology, 186(11), 3837-3847.
- Fothergill, J. L. (2020). Antimicrobial susceptibility testing for Pseudomonas aeruginosa: clinical implications of updated results. Clinical Microbiology Newsletter, 42(19), 161-166.
- Mittal, M., Siddiqui, M. Z., Bajpai, V., & Singh, P. (2021). Molecular techniques for identification of Pseudomonas aeruginosa: current status and future prospects. Journal of Applied Microbiology, 131(1), 28-44.
- Winstanley, C., O’Brien, S., & Brockhurst, M. A. (2016). Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends in Microbiology, 24(5), 327-337.
- https://www.ncbi.nlm.nih.gov/books/NBK557831/