TRANSCRIPTION
The process of formation of a messenger RNA molecule using a DNA molecule as a template is known as transcription.
Also, read Transcription in prokaryotes
TEMPLATE
The RNA is synthesized from a single strand of DNA that is known as the template of a DNA molecule.
TRANSCRIPTIONAL UNIT
The stretch of DNA that is transcribed into an RNA molecule is called a Transcriptional unit. A Transaction unit codes the sequence that is translated into protein.
ENZYMES INVOLVED IN EUKARYOTIC TRANSCRIPTION
The process of eukaryotic transcription is different in several ways from bacterial transcription. Eukaryotic transcription involves three major RNA polymerases:
- RNA polymerase I
- RNA polymerase II
- RNA polymerase III
| ENZYME | LOCATION | PRODUCT |
| RNA polymerase I | Nucleolus | Larger subunit of ribosomal RNA (rRNA) 28S, 18S, 5.8S |
| RNA polymerase II | Nucleus | Messenger RNA (mRNA), small nuclear RNAs (snRNAs), small interfering RNA (siRNA), microRNA (miRNA) |
| RNA polymerase III | Nucleus | tRNA |
FUNCTIONS OF ENZYMES
RNA polymerase I synthesize rRNA.RNA polymerase II is involved in mRNA synthesis. RNA polymerase III synthesizes tRNA. RNA polymerase II is a large molecule, that has a size of at least 500,000 Daltons, with about 10 or more subunits. The DNA entering is located in a clamp that closes down on polymerase II. A magnesium ion is also located on the active site. The DNA-RNA hybrid of nine base pairs in the transcription bubble is bound in a cleft formed by the two large polymerase subunits. The newly synthesized RNA leaves the polymerase beneath the lid region. The nucleoside triphosphate reaches the active site through a pore in the complex. RNA polymerase II needs many transcription factors to recognize its promoters, unlike bacterial polymerase.
They have combinations of several elements. Three of the most common are the TATA box (located about 30 base pairs upstream of the start point) and the GC and CAAT boxes (located between 50 to 100 base pairs upstream of the start site).
STEPS OF TRANSCRIPTION IN EUKARYOTES
- Initiation
- Elongation
- Termination
INITIATION
FACTORS ASSOCIATED WITH RNA POLYMERASE II
RNA polymerase II is associated with six transcriptional factors as follows:
- TFII-A
- TFII-B
- TFII-D
- TFII-E
- TFII-F
- TFII-H
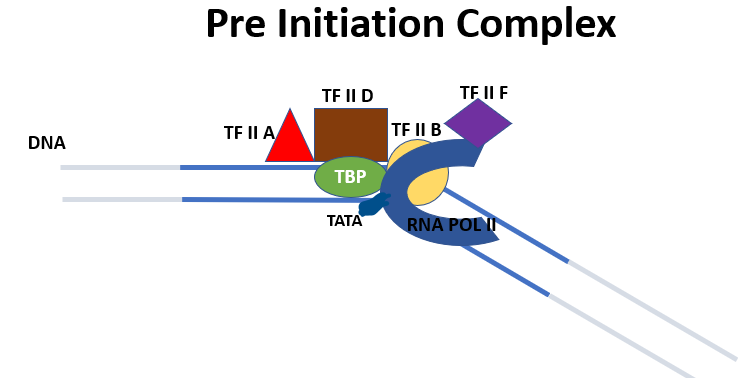
Pre-initiation complex
TF II D binds with TATA Box (30 nucleotides Upstream) in the promoter region. The promoter region is 40 nucleotides long (located upstream and downstream of the transcription start site). TF II D has a protein TBP (TATA-binding protein) that binds to the TATA sequence. It bends DNA by 80 degrees. TF II A binds before TF II D helps in stabilizing the binding of TF II D with the promoter. TF II B interacts with TBP and promoter region downstream of the TATA sequence. TF II B helps in the recruitment of RNA polymerase II.
RNA polymerase II cannot bind to the promoter on its own. TF II F helps RNA polymerase II to bind the promoter region. TF II F interacts with TBP and TF II B while recruiting the RNA polymerase. TF II F also prevents RNA polymerase to contact DNA outside the promoter. This is known as the pre-initiation complex.
Open Complex
Now TF II E binds pre initiation complex. TF II E helps in binding TF II H. TF II H is a large complex having 9 subunits. 2 subunits have ATPase activity they use energy from ATP and act like a helicase and melt the promoter. This causes a transition from pre initiation complex to open complex. 7 subunits have kinase activity that phosphorylates the C terminal domain or tail of RNA polymerase II leading to transcription elongation.

ELONGATION
Elongation factors
- TF E B
- TF II S
Non-coding strand is used as a template. As eukaryotic transcription proceeds, RNA polymerase traverses the complementary strand and uses base pairing complimentary to the DNA template to make an RNA copy. The template used is 3’ to 5’ direction and the RNA formed is 5’ to 3’ direction (exact copy of coding strand).
TF II H will use its helicase activity to unwind DNA. This requires energy from ATP hydrolysis. RNA pol II uses nucleoside triphosphate to synthesize RNA transcript. The Carboxyl terminal domain (CTD) is critical for elongation and it has to be phosphorylated during elongation. It contains many proline and serine residues.
TF II H has 9 residues 2 have ATPase activity remaining 7 subunits of TF II H have kinase activity that phosphorylates the CTD. This leads to promoter escape and transcription elongation proceeds by polymerase II. Factors that help in elongation are elongation factors.
FUNCTIONS OF ELONGATION FACTORS
TF E B: Kinase protein that phosphorylates serine residues at CTD of polymerase. This phosphorylation stimulates elongation
THE RATE OF TRANSCRIPTION IS NOT THE SAME IN EVERY REGION OF DNA. IT IS SLOW IN MANY REGIONS.
TF II S: helps to increase the rate of transcription at the region where the rate of transcription becomes slow.
As RNA pol II starts elongation, mRNA formation starts. mRNA formation occurs from 5’ to 3’ direction.
5’ CAPPING
Terminal gamma phosphate of nucleotide is removed by RNA triphosphatase from 5’. Geranyl transferase carries out a reaction between β phosphate of N terminal and α phosphate to form G-P-P-P. Then methyl group will attach to this G-P-P-P. this 5’ capping helps RNA to recruit on ribosomes to start the translation.

TERMINATION
TERMINATION FACTORS
- CSTF
- CPSF
Polyadenylation occurs at the 3՚ end of the mRNA. Two protein complexes are carried by the CTD of polymerase
1. CPSF (Cleavage and Polyadenylation Specificity Factor)
2. CSTF (Cleavage Stimulation Factor)
As the poly-a signals are transcribed by polymerase II into RNA, it will trigger the transfer of these factors to the RNA. CPSF and CSTF bound to the RNA, and recruit the endonuclease that removes the CSTF and cleaves the RNA right after the poly-a signal and becomes free.
POLYADENYLATION
Polyadenylation is mediated by:
- Poly-a polymerase (PAP) uses ATP as a precursor and adds the nucleotides (multiple stretches of ‘A’ nucleotides).
- Poly-a binding protein (PBP), binds with multiple stretches of ‘A’ nucleotides and prevents the formation of a secondary structure that leads to the formation of miRNA.
Cleavage of messenger RNA causes conformational changes in polymerase 2, and polymerase detaches itself from a template.
REFERENCES
Willey, J. M., Sandman, K. M., Wood, D. H., & Prescott, L. M. (2019). Prescott’s microbiology (11th ed.). McGraw Hill.