Definition
The process of conversion of double stranded DNA molecule into two identical DNA molecules is known as DNA replication. DNA replication in prokaryotes is different from DNA replication in Eukaryotes.
The double stranded DNA molecule is called a parental molecule and identical copies of DNA are known as daughter molecules. In a double helical DNA molecule, the nitrogenous bases are complementary along two strands so one of the DNA strands can act as a template strand for the synthesis of another strand.
Read Also Prokaryotes Vs Eukaryotes
Requirments in DNA replication
Many cellular proteins are required in the DNA replication of prokaryotes that carry out important and specific sequences of events during the replication process. Many enzymes are also involved in DNA replication. These enzymes are as follows:
| ENZYMES | FUNCTIONS |
| Helicase | It unwinds the double stranded DNA molecule |
| DNA polymerase | It synthesizes DNA and also helps in proofreading DNA and its repairment |
| Ligase | It joins DNA strands by forming a covalent bond between them, it joins the Okazaki fragments |
| Gyrase | It relaxes supercoiling ahead replication fork |
| Endo nuclease | It cuts DNA backbone and helps in repairment |
| Exo nuclease | It cuts DNA at an exposed end and also helps in repairment |
| RNA polymerase | It forms a copy of RNA from DNA |
| RNA primase | It is an RNA polymerase that forms RNA primers from DNA template |
| Topoisomerase | It separates the DNA circle at the end of DNA replication |
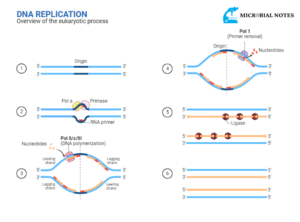
Replication process
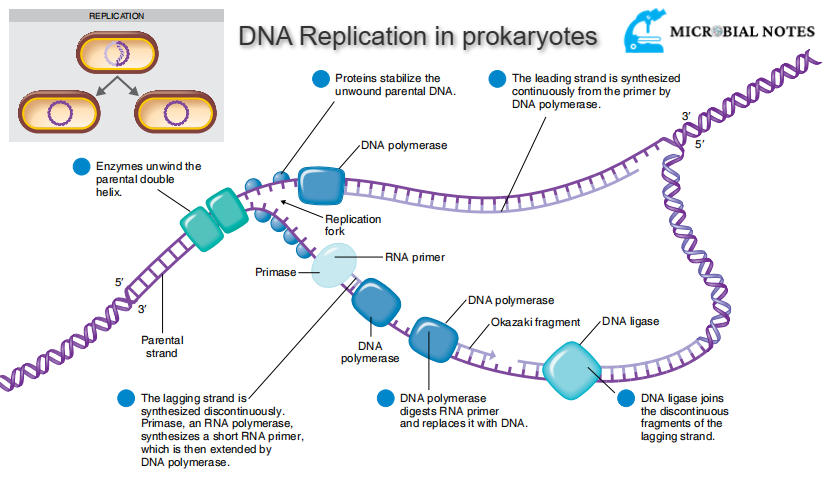
Unwinding of DNA molecule
When the DNA replication in prokaryotes starts, gyrase relaxes the supercoiling, and both strands of DNA are unwound by the helicase enzyme. Both strands get separated from each other forming two single stands of parental DNA. The nucleotides present freely in cytoplasm match up to the exposed bases of single stranded DNA parental strand i.e., cytosine will fit into the place where guanine is present in the parental DNA strand and adenine will fit into the place where thymine is present in the parental DNA strand.
This breaking of bonds and addition of new nucleotides is as follows:
- Parental DNA double helix gets separated by breaking weak hydrogen bonds between nucleotides on opposite strands with the help of a replication enzyme.
- Complementary nucleotides and each parental template strand form a hydrogen bond between them to make new base pairs.
- The formation of sugar phosphate bonds between sequential nucleotides on each daughter strand is catalyzed by the enzymes.
Nucleotide attachment to new molceule
The mismatched or improperly paired bases are removed and replaced by the replication enzymes. After the alignment, the newly placed nucleotide is joined to the growing DNA strand with the help of the DNA polymerase enzyme. Then for the addition of the next nucleotide, the parental DNA strand unwound a bit further. The replication fork is the point at which the replication occurs.
With the movement of the replication fork along the parental strand, each of the unwound parental single DNA strands adds new nucleotides. The parental or original strands and newly formed daughter strands are rewound. As each newly formed double stranded DNA molecule has one conserved (original/parental) strand and one new strand; the replication process is known as semi conservative replication.
Origin of replication:
A replicon is a DNA segment that undergoes replication. E. coli has a single replicon on its chromosomes, as do most prokaryotes. Replication is initiated at the origin of replication called Ori C which is 245 base pairs long and rich in AT sequences.
The structure of a DNA molecule is such that the DNA strand’s orientation is in opposite directions relative to each other. The carbon of the sugar component of every nucleotide is numbered 1’ to 5’ and the sugar components of one strand are upside down relative to the other. At the 3’ carbon end, hydroxyl is attached and it is known as the 3’ end of the DNA strand. At 5’ carbon phosphate is attached and it is known as the 5’ end of the DNA strand. The direction of one strand is 5’ to 3’ while the direction of another strand is 3’ to 5’.
Leading and lagging strands
The strand (5’ to 3’) which is complementary to the 3’ to 5’ parental DNA strand is synthesized continuously towards the replication fork because the polymerase can add nucleotides in this direction. This continuous strand is known as the leading strand. The other strand complementary to 5’ to 3’ parental DNA is extended away from the replication fork in small segments known as Okazaki fragments. The strand with Okazaki fragments is known as the lagging strand.
DNA polymerase can add nucleotides at only the 3’ end of the DNA strand so, with the movement of the replication fork along parental DNA, both new strands grow in different directions. A vast amount of energy is required in the process of DNA replication. Nucleotides (nucleoside triphosphate) provide energy for the process.
Replication fork events are as follows:
- Parental double helix DNA molecule structure is unwound by the enzymes.
- This unwound structure is stabilized by the proteins.
- DNA polymerase continuously synthesizes the leading strand.
- The synthesis of the lagging strand is discontinuous. A short RNA primer is synthesized by primase and an RNA polymerase which is then extended by DNA polymerase.
- RNA primer is digested by DNA polymerase and replaced by DNA.
- The discontinuous fragments of the lagging strand are joined by DNA ligase.
Bidirectional replication
Some bacteria like E. coli do DNA replication bidirectionally around the chromosome. Both replication forks move in the opposite direction away from the origin of replication. As the bacterial chromosome is circular (a closed loop) both replication forks meet after replication. These loops are then separated by an enzyme known as topoisomerase.
Each copy of the origin binds to the membrane at opposite poles after duplication hence each daughter cell receives a copy of the DNA molecule. this is the formation of one complete chromosome.
Proofreading:
DNA polymerases III have the ability to proofread by using 3’ → 5’ exonuclease activity. Is important for the survival of an organism that its DNA must be replicated without any error. Mismatched bases or errors led to mutation. If any mismatch nucleotide is detected, it will be removed and replaced by the accurate nucleotides.
References
Tortora, G. J., Funke, B. R., & Case, C. L. (2021). Microbiology: An introduction. Pearson Education Limited.