Types of PCR (Polymerase chain reaction)
PCR is a technique used to replicate DNA in in-vitro conditions. There are many types of PCR mentioned below
Also Read PCR (Polymerase Chain Reaction): Its principle, components, and applications
Amplified fragment length polymorphism(AFLP)PCR:
- It is a PCR-based method that creates distinctive fingerprints for target genomes by selectively amplifying a portion of digested DNA fragments.
- Using this method, any organism may swiftly produce a huge number of marker fragments without needing to know the genetic sequence beforehand.
- AFLP PCR breaks down genomic DNA using restriction enzymes and enables the attachment of adaptors to the sticky ends of the fragments.
- The adaptor sequence-complementary primers are then used to choose a portion of the restriction fragments to be amplified.
- The amplified sequences are isolated and made visible using agarose gel electrophoresis after denaturing.
- Applications for AFLP PCR include assessing genetic diversity within or among closely related species, inferring population-level phylogenies and bio-geographic patterns, creating genetic maps, and determining cultivar relatedness.
Allele-specific PCR:
- Single nucleotide polymorphism can be examined using the allele-specific polymerase chain reaction (AS-PCR) method, which is based on allele-specific primers.
- Amplification Refractory Mutation System (ARMS-PCR) is another name for allele-specific PCR, which refers to the employment of two separate primers for two different alleles.
- The first set of primers is the mutant set, which is resistant to the normal PCR, and the second set of primers is the normal set, which is resistant to the mutant PCR response.
- These primers’ 3′ ends have been changed so that some primer sets can amplify the mutant allele while others can amplify the normal allele.
- A single allele can be amplified by the primer thanks to this mismatch.
- It is frequently used to identify single gene point mutations in diseases like thalassemia and sickle cell anemia.
- Additionally, it is utilized to directly determine the genotype of ABO blood groups.
Alu PCR:
- Alu PCR is a quick and simple DNA fingerprinting method that examines numerous genomic loci simultaneously that are surrounded by Alu repeating elements.
- Short DNA segments known as Alu elements were first identified by the Arthrobacter luteus (Alu) restriction endonuclease.
- One of the most prevalent transposable elements in the human genome, all elements also play a role in the evolution and have been employed as genetic markers.
- In Alu PCR, the PCR is carried out using two fluorochrome-labeled primers that are complementary to those sequences, and the PCR products are then examined by
- Alu insertions have been used to treat different cancer types and genetically inherited human disorders. Therefore, this PCR is crucial in the identification of various illnesses and mutations.
Assembly PCR:
- Using many shorter fragments, big DNA oligonucleotides can be assembled using the assembly PCR technique.
- Oligonucleotides utilized in PCR are 18 base pairs in length, although PCR lengths up to 50 bp are employed in assembly to ensure proper hybridization.
- The oligonucleotides bind to complimentary fragments during the PCR cycles, and the polymerase enzyme subsequently fills in the gaps.
- As a result, depending on which oligonucleotides bind to one another, different segments randomly lengthen during each cycle of this PCR.
- Assembly PCR is used to increase protein yield and can also be used to generate significant amounts of RNA for structural or biochemical research.
Asymmetric PCR:
- An adaptation of the PCR method known as asymmetric PCR is used to preferentially amplify one strand of the original DNA over the other.
- The excessive number of primers for a selected strand distinguishes asymmetric PCR from conventional PCR.
- The lower concentration limiting primer is quantitatively integrated into freshly manufactured double-stranded DNA and used up as the asymmetric PCR proceeds.
- As a result, following the depletion of the limiting primer, linear synthesis of the targeted single DNA strand is produced from the surplus primer.
- When only one of the two complementary strands needs to be amplified, like in sequencing and hybridization probing, it is helpful.
COLD PCR:
- Unaffected by the type of mutation or where it is located on the amplicon, the co-amplification at lower denaturation temperature-based polymerase chain reaction (COLD-PCR) selectively amplifies low-abundance DNA variants from mixtures of wild-type and mutant-containing (or variant-containing) sequences.
- This approach is based on changing the critical temperature at which DNA with mutations melts more quickly than DNA without mutations.
- After denaturation, there is an intermediate annealing process that permits the hybridization of the wild-type and mutant alleles. The melting temperature of the ds DNA is somewhat changed by this mismatch.
- The melting of these hetero-duplexes will serve as a model. As a result, a larger percentage of the minor variant DNA will be amplified and available for use in subsequent PCR rounds.
- Particularly in diverse tumors and body fluids, PCR is crucial in the detection of mutations in oncology material.
- This PCR also aids in disease staging, molecular profiling for prognosis, and individualized therapy planning as well as the detection of residual illness following surgery or chemotherapy.
Colony PCR:
- Colony PCR is a technique that uses specially designed primers for the inserted DNA to identify the DNA of interest that has been introduced into the plasmid.
- Using two sets of primers, the bacterial colony harboring the plasmid can be amplified directly.
- The first set of primers amplifies the insertion sequence; the second set of primers amplifies plasmid DNA other than the inserted DNA and is specific to the vector.
- The master mix, which also contains all the other PCR reagents, is immediately supplemented with a bacterial colony.
- Colony PCR’s primary use is in determining the proper ligation and incorporation of inserted DNA into bacteria and yeast plasmids.
Conventional PCR:
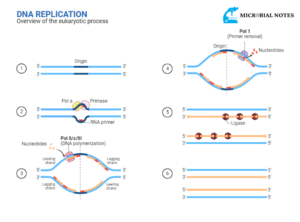
- Using a test tube technology called the polymerase chain reaction (PCR), it is possible to selectively amplify a “target” DNA sequence millions of times in a matter of hours.
- A DNA-polymerase enzyme, which participates in the replication of the cellular genetic material, is used in PCR to enable the synthesis of particular DNA fragments.
- As a tiny fragment (primer) is joined to one of the DNA strands in the exact site selected to start the synthesis, this enzyme creates a complementary sequence of DNA.
- A specific DNA sequence is amplified with billions of copies when primers restrict the sequence that can be reproduced.
- Conventional PCR is used in forensic investigations, medical and diagnostic procedures, infectious illness detection, selective DNA isolation, DNA amplification, and DNA quantification.
Digital PCR(dPCR):
- The quantitative PCR technique known as digital PCR (dPCR) offers a sensitive and effective method for determining how much DNA or RNA is present in a sample.
- Prior to the amplification phase in dPCR, the initial sample mix is split up into a large number of separate wells, which determines if the target sequence is present in each well or not.
- The absolute number of targets present in the original sample is calculated based on the presence or absence of fluorescence in the amplified reaction wells.
- Wells having a fluorescent signal are positive and given a score of “1,” whereas wells without one are considered negative and given a score of “0.”
- The Poisson statistical analysis is then used to calculate the concentration of the target sequence that was present in the first sample.
- When a well-calibrated standard is not available, dPCR is typically used to estimate the total quantities of DNA and RNA viruses, bacteria, and parasites in a range of clinical specimens.
Fast cycling PCR:
- Fast cycling PCR is a PCR-based method that allows for much shorter cycle times during the amplification of particular PCR products.
- The only difference between this technique and traditional PCR is the timing of the amplification.
- By increasing the Taq DNA polymerases’ affinity for small single-stranded DNA fragments, the buffer utilized in this PCR may successfully anneal primers in about 5 seconds.
- For processes that require quick cycles, fast-cycling PCR is crucial. It also aids in the speedy identification of illnesses and mutations.
High-Fidelity PCR:
- High-fidelity PCR, a modified version of PCR, uses a DNA polymerase with a low error rate and produces highly accurate copies of the DNA of interest.
- During amplification, these enzymes have a high affinity for the proper nucleoside triphosphate.
- The design of the active site complex causes incorporation to be hindered in the event of an erroneous binding in the polymerase active site.
- For research like cloning, SNP analysis, and NGS applications where the outcome depends on the proper DNA sequence, high-fidelity amplification is crucial.
High-Resolution Melt (HRM) PCR:
- It is a remarkably effective method for identifying polymorphisms, mutations, and epigenetic variations in double-stranded DNA samples.
- Compared to other genotype technologies like sequencing and Taqman SNP typing, it is incredibly cost-effective. It is therefore perfect for extensive genotype operations.
- It can accurately genotype a large number of samples quickly since it is quick and powerful.
- It is easy. In any laboratory with access to a real-time PCR machine that is HRM competent, powerful genotyping can be carried out by non-geneticists using a high-quality HRM assay.
- Hot Start PCR:
A unique variation on the standard polymerase chain reaction (PCR), hot start PCR minimizes the production of undesirable products and the development of primer-dimers as a result of non-specific DNA amplification at room temperature. - The fundamental idea behind hot-start PCR is the removal of one or more reagents from the reaction mixture before heating it to the denaturation temperature.
- Hot start PCR greatly decreases non-specific binding, prevents primer dimers from forming, and frequently boosts product yields. Additionally, it needs less work and lowers the possibility of contamination.
In-Situ PCR:
- Using frozen or paraffin-embedded cells or tissue sections, in-situ polymerase chain reaction (In-situ PCR) is an efficient approach for detecting minute amounts of uncommon nucleic acid sequences in order to compartmentalize such sequences within the cells.
- With this technique, tissue is fixed to preserve the cell shape before being treated with proteolytic enzymes to create a channel for the PCR reagents to interact with the target DNA.
- The reagents amplify the target sequences, which are subsequently identified using conventional immunocytochemical procedures.
- The diagnosis of infectious diseases, the measurement of DNA, the detection of even minute amounts of DNA, and the study of organogenesis and embryogenesis are all possible with in-situ PCR.
Intersequence-specific (ISS)PCR:
- InterSequence-Certain PCR, also known as ISSR-PCR, is a technique for creating unique DNA fingerprints by using primers chosen from specific sequences repeated throughout a genome.
- The method amplifies primarily the inter-SSR sequences of various sizes by using microsatellites, which are typically 16–25 bp long, as primers in a single primer PCR reaction that targets several genomic loci.
- Genomic fingerprinting, genetic diversity and phylogenetic research, genome mapping, and gene tagging are all applications of ISSR PCR.
Inverse PCR:
- One of the numerous polymerase chain reaction variations used to amplify DNA when only one sequence is known is the inverse polymerase chain reaction (Inverse PCR).
- Inverse PCR allows amplification to occur even if only one sequence is available from which primers may be constructed, in contrast to conventional PCR which requires primers complementary to both terminals of the target DNA.
- The inverse PCR uses several rounds of restriction digestion followed by ligation to create a looping fragment that may be primed for PCR using just one segment of a predetermined sequence.
- The temperature-sensitive DNA polymerase then amplifies the DNA, similar to other polymerase chain reaction procedures.
- For pinpointing the sites of numerous transposes and retroviruses in the host DNA, inverse PCR is very helpful.
LATE ( Linear-After-The-Exponential) PCR:
- Asymmetric PCR is modified by LATE (Linear-After-The-Exponential) PCR, which employs a limiting primer with a higher melting temperature than the excess primer to ensure reaction efficiency as the limiting primer concentration falls during the reaction.
- The exponential phase of LATE-PCR starts with an amplification efficiency that is comparable to that of traditional PCR. The reaction quickly changes to linear amplification once the limiting primer is exhausted, and the single-stranded product is maintained for numerous extra heat cycles.
Ligation-mediated PCR:
- A modified version of traditional PCR known as “ligation-mediated PCR” is made possible by starting with only one end known and adding the second end by tying a special DNA linker.
- Small DNA fragments known as “linkers” (or adaptors) are used in ligation-mediated PCR and are first joined to portions of the target DNA.
- The target fragments are then amplified using PCR primers created to bind to the linker regions.
- For DNA sequencing, genome walking, and DNA footprinting, this technique is used.
Long-Range PCR:
- Long-Range PCR is a technique for amplifying longer DNA fragments that are generally difficult or impossible to do so using standard PCR techniques or reagents.
- By modifying high-efficiency polymerases with improved DNA binding, lengthy fragments can be amplified with great processivity and accuracy utilizing long-range PCR.
- Using less time and money, this technique enables the amplification of more extensive targets.
Methylation-specific PCR(MSP):
- A technique for identifying and examining DNA methylation patterns in CpG islands is called methylation-specific PCR (MSP).
- Two primer pairs, each of which is detectably methylated and unmethylated DNA, are used in PCR to modify DNA and perform MSP.
- After the DNA has been treated with bisulfite to turn the cytosine into uracil, the methylated regions are then preferentially amplified using primers designed for
- Since excessive methylation of CpG dinucleotides in the promoter represses the production of genes, it is crucial to find methylated patterns.
Miniprimer PCR:
- Miniprimer PCR is a novel PCR technique that utilizes a designed polymerase and 10-nucleotide “mini primers”.
- New 16S rRNA gene sequences that would not have been discovered using conventional primers are found to be revealed by this technique.
- A thermostable polymerase enzyme is used in mini primer PCR and can extend from brief primers (9 or 10 nucleotides).
- The 16S (or eukaryotic 18S) rRNA gene is amplified using this technique, which enables PCR targeting to smaller primer binding areas and is utilized to amplify highly conserved DNA sequences.
Multiplex PCR:
- Molecular biology methods like multiplex PCR are frequently employed to amplify many targets simultaneously.
- In multiplex PCR, DNA is amplified in a thermal cycler using several primers and a temperature-mediated DNA polymerase.
- It is necessary to tune each primer pair created for multiplex PCR such that each pair’s annealing temperature during PCR is the same.
- When several sequences are targeted simultaneously, more information can be obtained from a single test run than would otherwise be possible without using a lot of reagents or exerting a lot of time and energy.
- Numerous fields, including genotyping, mutation and polymorphism analysis, microsatellite STR analysis, the detection of infections or genetically modified organisms, etc., have used this technology.
- Multiplex PCR is used in diagnostic labs to identify various bacteria that produce the same kinds of diseases.
Nanoparticle-Assisted PCR(nanoPCR):
- Small molecules with certain physical properties that improve the reaction are included in a PCR using nanoparticles.
- According to one theory regarding gold nanoparticles, these particles adsorb some of the polymerases and regulate the quantity of polymerase that is still present in the system, which may be important to improve the specificity of the reaction.
- According to a different explanation, they adsorb primer pairs and reduce the melting temperature during the creation of duplexes between properly paired and imperfectly paired primers, increasing the specificity of the reaction.
- High sensitivity, high specificity, and high selectivity are advantages of nanoparticle-associated PCR, which has been extensively employed in gene sequencing and viral detection.
Nested PCR:
- The specificity of the reaction is increased by preventing non-specific binding with the aid of two sets of primers in nested PCR, a helpful modification of the PCR technology.
- While another set of primers binds directly at the target region, the first set of primers amplifies bigger fragments by binding outside of our target DNA.
- A second set of primers amplifies only the target DNA in the subsequent round of amplification.
- For phylogenetic analyses and the detection of various diseases, nested PCR is a useful technique.
- The method is more sensitive, so even if the sample has less DNA, it can still be amplified, which is not possible with the traditional PCR method.
Overlap extension PCR(OE-PCR):
- The term “Splicing by Overlap Extension” or SOEing is another name for this technique.
- Cross-over extension PCR is an effective method that is frequently used for cloning huge complicated fragments, editing cloned genes, and joining two gene components.
- Long DNA segments are formed by combining shorter ones.
- It is employed for numerous site-directed big fragment insertion, deletion, and replacement operations as well as efficient gene cloning.
- Site-directed mutagenesis, the production of chimeric molecules, and even the cloning of lengthy gene segments by joining smaller sections have all been demonstrated to be effective uses for it.
Real-Time PCR (Quantitative PCR (qPCR)):
- Amplification of a target DNA sequence is combined with the measurement of the concentration of that DNA species in the reaction in a process known as quantitative PCR (qPCR), also known as real-time PCR or quantitative real-time PCR.
- The analysis of the PCR results by gel electrophoresis takes time in conventional PCR. By allowing for the real-time detection of products during the exponential phase, qPCR simplifies the analysis.
- The use of fluorescent dye is essential to the real-time PCR method.
- Using the fluorescent dye or fluorescently labeled oligonucleotides, the concentration of nucleic acid contained in the sample is measured.
- Pathogen genotyping and quantification, microRNA analysis, cancer diagnosis, microbial load assessment, and the detection of GMOs all use q-PCR.
Repetitive sequence-based PCR:
- Rep-PCR, a modified form of PCR, uses primers that specifically target noncoding repetitive sequences scattered across the bacterial genome.
- Such clusters of non-coding, repeating sequences can operate as several genetic targets for oligonucleotide probes, allowing the creation of distinctive DNA profiles or bacterial strain fingerprints.
- Rep-PCR is mostly used to molecularly type distinct bacterial strains. It is also used to distinguish different pathogens epidemiologically.
Reverse Transcriptase PCR(RT-PCR):
- Reverse transcription PCR (RT-PCR) is a variation of traditional PCR in which complementary DNA (cDNA) molecules are first created from RNA molecules before being amplified by PCR.
- Reverse transcriptase is used in RT-PCR to first turn the RNA template into a complementary DNA (cDNA). Then, using PCR, the cDNA serves as a template for exponential amplification.
- One tube can be used for RT-PCR, or two tubes can be used in two separate steps. With fewer opportunities for contamination and variation assimilation, the one-step technique is more efficient.
- Research methodologies, gene insertion, genetic illness diagnostics, and cancer detection all use RT-PCR.
Reverse-Transcriptase Real-Time PCR(RT-qPCR):
- Reverse Transcriptase Real-Time PCR, which RT-PCR and q-PCR frequently create (RT-qPCR).
- This enables real-time DNA measurement following amplification.
RNase H-dependent PCR:
- The primers for the RNase H-dependent PCR have a detachable amplification block at the 3′ end.
- Only an RNase enzyme cleavage activity upon hybridization to the corresponding target sequence will allow the blocked primer to amplify.
- Since the RNase H enzyme exhibits very little enzymatic activity at low temperatures, a hot start can be achieved without changing the DNA polymerase.
- Similarly to this, the enzyme’s cleavage effectiveness is decreased when mismatches are present close to the RNA residue.
- As a result, the RNase H enzyme’s activity reduces the creation of primer dimers and non-specific binding, which facilitates efficient hybridization.
Single Specific Primer PCR:
- SSP-PCR, a PCR-based method, allows for the amplification of genes for which only a small amount of incomplete sequence information is available.
- It enables unidirectional genome wandering from well-known into uncharted chromosomal regions.
Single Specific Primer-PCR (SSP-PCR):
- As a result, double-stranded DNA can be amplified even when the sequence information is only present at one end.
- This technique, known as single specific Primer-PCR (SSP-PCR), enables unidirectional genome walking from known into unknown areas of the chromosome and the amplification of genes for which only partial sequence information is available.
Solid Phase PCR:
- A special PCR method called solid-phase PCR (SP-PCR) enables target nucleic acid amplification on solid supports with one or both primers bound on the surface.
- The primer-dimer formation is prevented by the spatial separation of the primers, which also enables better multiplexing amplification. The primer-dimer formation is greatly reduced.
- This unique method’s main concept is to connect the primers’ 5′ ends to a surface rather than allowing them to freely disperse in a bulk solution.
- The surface of a freely diffusing DNA target can be grabbed, and the polymerase can then copy it.
- After the annealing stage, the original DNA molecule returns to the solution while the copy remains affixed to the surface.
- The amplification process can begin when the free end of the attached copy hybridizes with the primer (attached to the surface) that complements its sequence.
Suicide PCR:
- Suicide PCR is frequently employed in research where preventing false positives and making sure the amplified fragment is particular are of utmost importance.
- According to the approach, each primer pair should only be used once during a PCR and should not have been used during any positive control PCR reactions.
- Always choose a genomic area for these primers that have never been amplified with any other set of primers or this specific primer.
- This setup makes sure that there isn’t any contaminated DNA from earlier PCR reactions in the lab, which may otherwise result in false positives.
- Paleogenetics research, which looks at genetic material that has been conserved from the remains of ancient species, employs suicide PCR.
Thermal asymmetric interlaced PCR (TAIL-PCR):
- TAIL PCR is an effective method for recovering DNA fragments close to recognized sequences.
- TAIL -PCR employs three nested primers in a series of reactions together with an arbitrary degenerate primer with a low melting temperature to allow for the thermal regulation of the relative amplification frequencies of particular and non-specific products.
- The TAIL-PCR products that have not been purified can be immediately sequenced thanks to the method’s high degree of accuracy.
- Additionally, it enables the cloning of fully functional genes.
Touch Down PCR:
- Touch Down PCR is a variation of PCR in which the starting annealing temperature is higher than the primer’s ideal Tm and is progressively lowered over succeeding cycles until the Tm temperature, or “touchdown temperature,” is reached.
- Touchdown PCR lowers the annealing temperature toward the end of the reaction to maximize efficiency while increasing specificity at higher temperatures.
A variable number of Tandem Repeats (VNTR) PCR:
- They serve as significant forensic scientific identifiers for the individual.
- In VNTR PCR, segments are produced that show variations across species but little variation within a species.
- The polymerase chain reaction (PCR) can successfully amplify from a very little amount of genomic DNA (PCR).
- The PCR-based variable-number tandem repeat (VNTR) analysis stood out among the genotyping methods as a viable approach for typing M. tuberculosis.
What are the uses of PCR?
The COVID-19 epidemic has made many individuals more accustomed to the term “PCR test.” However, the method has a lot more medical applications besides detecting COVID-19Trusted Sources.
A PCR test can also be used by medical professionals to find minute quantities of cancer cells and genetic alterations that can lead to illness. PCR tests can also identify additional organisms that might cause illnesses, including:
- Influenza
- Tuberculosis reliable source
- Dependable source of HIV/Ebola
- Reputable source for hepatitis C
What is the Summary of PCR?
A frequent study method is PCR testing. It can aid in the detection of genetic alterations, malignant cells, or pathogens like SARS-CoV-2 in a medical setting.
A sample of bodily fluid is drawn for the test, and the genetic material is processed to create several duplicates of itself. A PCR test for COVID-19 can take longer than other tests to produce findings, but the results are frequently more accurate.